Here's the thing, inbreeding is the mating of pairs who are closely related, genetically. It can lead to homozygosity, which is when a cell has two identical alleles of the same gene on both homologous chromosomes. In the punnett square below, the individuals with BB and bb alleles are homozygous for that particular trait.
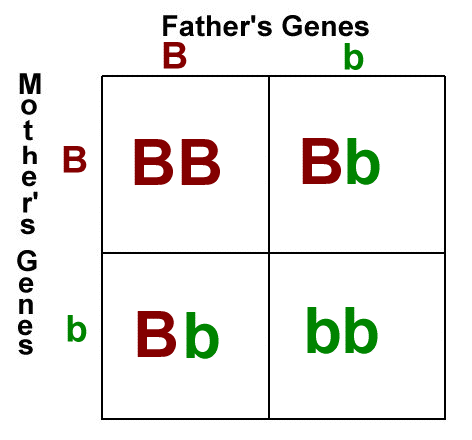
Homozygosity is not necessarily a bad thing. It's how some of us end up blonde - because blonde is a recessive trait (not dominant), and a person needs to be homozygous recessive for hair color to end up with blonde hair. A person heterozygous for hair color would carry one copy of the dominant allele (brown) and one copy of the recessive allele, but would express the dominant allele and be brunette. In reality, heredity is far more complex than this, what I've described here is a simplification of the process. A lot of genetic disorders are recessive though. That means that people can be carriers but not express a disorder. When two carriers have a child, the child has a chance of being homozygous recessive for that trait, since both parents carry a copy of the allele. It's the reason why purebred animals often have health problems.
Royal families have historically been inbred to maintain "pure" blood lines. Because of its isolation, the population of Iceland was also historically inbred. Inbreeding is never going to lead to mutant children with two heads, or something ridiculous like that, because humans do not carry genes for two heads. In fact, no animals do.
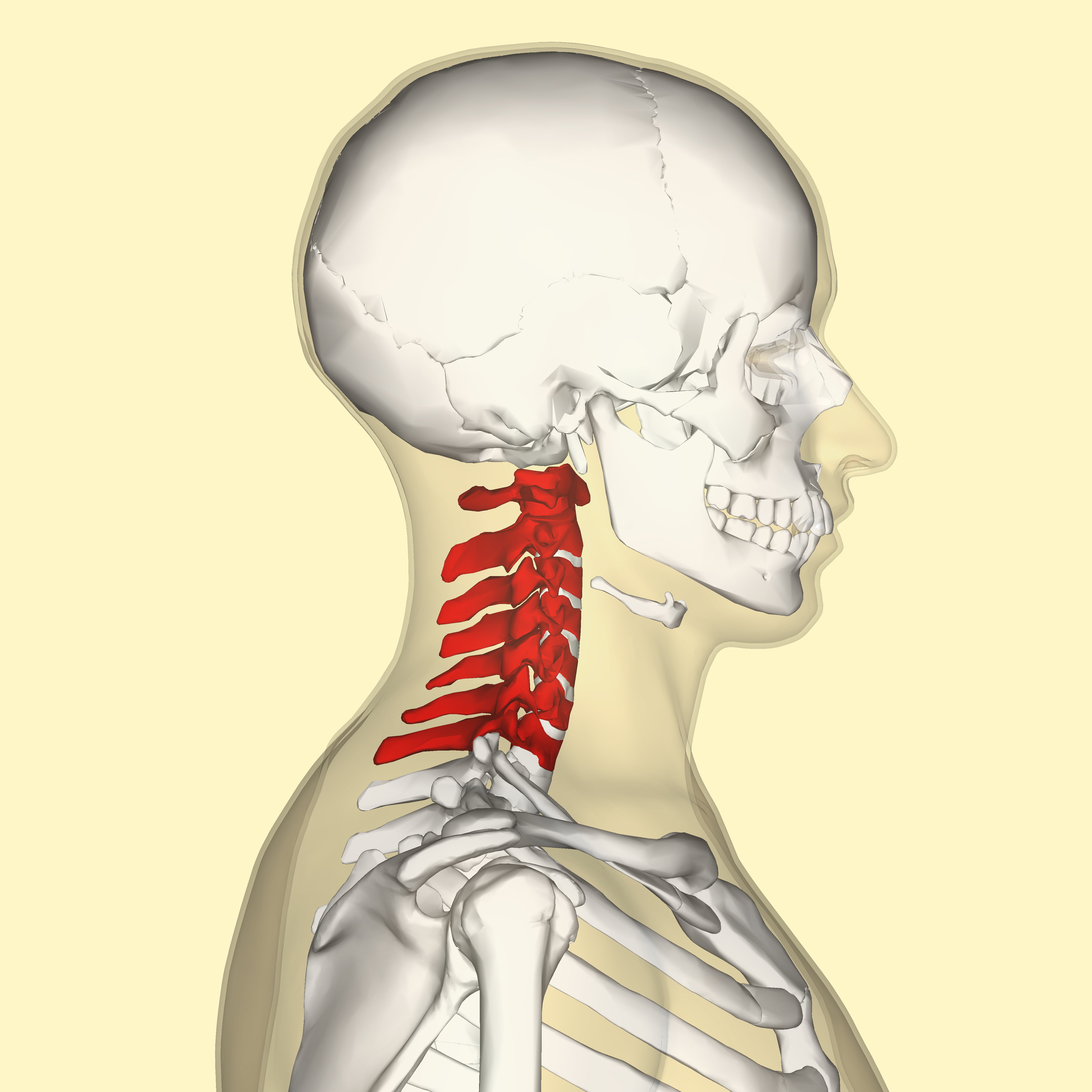 But inbreeding depression in a population can be a really serious issue. A new open access paper published this past week suggests that woolly mammoths (Mammuthus primigenius) may have inbred themselves into extinction. See, there's a strong conservation across vertebrate species for the number of cervical vertebrae, which is typically 7. There are interspecies variations, just like there are for any other trait, but intraspecies variation is limited. The most typical variation in vertebral number is the presence of ribs in the seventh vertebra.
But inbreeding depression in a population can be a really serious issue. A new open access paper published this past week suggests that woolly mammoths (Mammuthus primigenius) may have inbred themselves into extinction. See, there's a strong conservation across vertebrate species for the number of cervical vertebrae, which is typically 7. There are interspecies variations, just like there are for any other trait, but intraspecies variation is limited. The most typical variation in vertebral number is the presence of ribs in the seventh vertebra.It looks like this:

The presence of a cervical rib is, in itself, harmless. But still, 90% of people born with a cervical rib die before reaching reproductive age, thus presenting a huge selective pressure against cervical ribs. Cervical ribs are often associated with multiple major congenital abnormalities. The selective pressure against cervical ribs is thus a selective pressure against the other defects it is often associated with.
Three cervical vertebrae of the woolly mammoth were recently found in the North Sea during infrastructural work, and were donated to the Natural History Museum of Rotterdam. Two of those three cervical vertebrae contained facets of cervical ribs. The authors looked through other fossil records to find the prevalence of this trait in other mammoths, and in the closely related extant species of African and Asian elephants.
The authors found that 33% of samples from mammoths contained cervical ribs, compared to 3.6% for the two extant elephant species. This is a statistically significant finding. The authors then concluded that, with this high incidence of cervical ribs, coupled with the limited genetic diversity of the time, it is likely that inbreeding is what caused the development of these protrusions. As you can imagine, the internet went bananas over this.
But I have a serious problem with the methodology of this study. They looked at a total of 16 samples of mammoth cervical vertebrae, and 28 samples of the cervical vertebrae of extant elephants. This sample size is too small to be considered valid and to support the generalizations that were made. Furthermore, data was unavailable for 7 of the 16 samples from mammoths. So they found 3 cases in 9 samples (33%), and reported this as statistically significant. To be honest, I don't even understand why they would have reported that they looked at 16 samples, when they really could only measure 9 of them. This was flagged in the peer-review process. In fact, another group has recently been reported to have ruled out inbreeding among the dwindling mammoth population altogether. While I'm not going to deny the findings, I'll reiterate that I think the authors have jumped the gun in terms of generalizing their results, and I think that the way this study was reported has now spread a huge amount of misinformation regarding the extinction of woolly mammoths.