The highlights of day 2 are after the jump:
While the tone of yesterday's session focused on the speed of these new technologies in diagnosing disease and creating efficient and optimal treatment, today's sessions had more of an urgent tone. More specifically, the urgency in applying genomics technology to address important and pressing issues in climate change and the environmental impact on human health. The need for good communication in translating science into policy practice was also a common concern that arose throughout the day.
We spent the morning hearing about about epigenetics and epigenomics. Even when you know the genomic sequence of an individual, you can only predict the development of disease in 20% of the cases, so how do we account for the rest of it? The remainder is explained by epigenetic factors. You may remember some recent research from the lab of Andrew Feinberg regarding the epigenetic fingerprints left by cancer. Dr. Feinberg, an absolutely lovely man from Johns Hopkins University (who was pretty excited to learn of my previous article on his research), talked about the epigenetic factors associated with cancer, and how cancer develops from repeated damage-repair cycles. He has also developed the field of epigenetic epidemiology, which examines the environmental and genetic contributions to disease, in a combined epigenome-wide association study (EWAS)/GWAS method. With this technique, his group is finding promising results in the prediction of future disease development and which genes will be affected.
 We also heard from George Weinstock, from The Jackson Laboratory for Genomic Medicine, who spoke about the human microbiome, and how our bodies are essentially scaffolds for microbes. The microbiome contributes over 1M genes, far outnumbering the 23,000 genes found in the human genome. There is an evolutionary significance to the plethora of microbial species living in the human body - these microorganisms provide us with a number of functions that don't need to be encoded in our own genome. Dr. Weinstock's work has focused on metagenomic analysis of the human microbiome, a technique that sequences all the available nucleic acids in a given sample, without needing to culture. By comparing the microbiome of different groups of people - sick v. healthy, young v. elderly, etc., Dr. Weinstock has found significant differences in the microbial populations, enough to possibly predict disease outbreaks before they happen. His team has also been experimenting with finding "optimal" microbiomes by looking at the microbial compositions of atheletes.
We also heard from George Weinstock, from The Jackson Laboratory for Genomic Medicine, who spoke about the human microbiome, and how our bodies are essentially scaffolds for microbes. The microbiome contributes over 1M genes, far outnumbering the 23,000 genes found in the human genome. There is an evolutionary significance to the plethora of microbial species living in the human body - these microorganisms provide us with a number of functions that don't need to be encoded in our own genome. Dr. Weinstock's work has focused on metagenomic analysis of the human microbiome, a technique that sequences all the available nucleic acids in a given sample, without needing to culture. By comparing the microbiome of different groups of people - sick v. healthy, young v. elderly, etc., Dr. Weinstock has found significant differences in the microbial populations, enough to possibly predict disease outbreaks before they happen. His team has also been experimenting with finding "optimal" microbiomes by looking at the microbial compositions of atheletes.Art Petronis, from the Centre for Addiction and Mental Health (CAMH), spoke about epigenomics and psychiatric illness. We heard a bit about the discordance in twin studies yesterday from Guy Rouleau; identical twins (who share 100% of their genetic makeup) don't always end up with the same psychiatric disorders, which points toward an environmental component in mental illness. Luckily for us, the epigenome is far more plastic than the genome, and so we can avoid the development of mental illness with targeted policy interventions.
We also heard about the One Health initiative from John Harding of the University of Saskatchewan. One Health is a worldwide strategy aiming to improve human health by improving animal and ecosystem health. Our interconnectedness with our ecosystem determines so much about our health, as seen, for example, in the 1999 Nipah virus outbreak in Malaysia and the current Ebola outbreak in West Africa. These types of diseases are often transmitted to humans from accidental hosts, which include pigs, rodents, and sadly even dogs. Dr. Harding talked about the genomics research at the University of Saskatchewan, which is attempting to find solutions for Salmonella transmission in the Canadian pork industry. These solutions could are worth $9.3B, and 45,000 jobs.
 In the afternoon, we moved away from human health and focused more on the environment. Louis Fortier from the University of Laval spoke about the impact of climate change on the Arctic (spoiler: it's worse than for the rest of the Earth). His talk was definitely grim: if we continue on our current trends, we could be looking at a summer in which the Arctic is ice-free by 2030! Melting sea ice and glaciers are having a huge effect on infrastructure, and on the mental and physical health of Inuit and other northern populations in Canada. Dr. Fortier has studied the Arctic climate from the CFI-funded ice breaker Amundsen, and is combining his research findings with community consultations to make effective policy recommendations for this region.
In the afternoon, we moved away from human health and focused more on the environment. Louis Fortier from the University of Laval spoke about the impact of climate change on the Arctic (spoiler: it's worse than for the rest of the Earth). His talk was definitely grim: if we continue on our current trends, we could be looking at a summer in which the Arctic is ice-free by 2030! Melting sea ice and glaciers are having a huge effect on infrastructure, and on the mental and physical health of Inuit and other northern populations in Canada. Dr. Fortier has studied the Arctic climate from the CFI-funded ice breaker Amundsen, and is combining his research findings with community consultations to make effective policy recommendations for this region. Dr. Fortier mentioned that genomics studies are extremely useful in studying invasive species that are spreading as a result of climate change in the Arctic. This segued nicely into Paul Hebert's talk about planetary biodiversity. Dr. Hebert from the University of Guelph uses genomics to study biodiversity. He started his talk by showing us 6 species that were the cornerstone of his youth in Kingston, Ontario - species which the youth of Kingston today will never see, either because they are too rare, or because they are gone from the region (or altogether). Humans have always been to blame for large-scale extinctions. When we first moved out of Africa 25,000 years ago, we wrecked havoc on the indigenous species of our new habitats. Of course, now we're looking at a human cause of the next mass extinction, and it's not because we're eating these species, but because we're destroying their habitats and food/water supplies. Globalization has created a new Pangaea, which threatens endemic taxa by introducing a number of invasive species. Dr. Hebert suggests that using genomics in biosurveillance to identify any invasive species as soon as possible, and creating a DNA database of the genetic information of all the species on Earth could avoid another mass extinction. So far, his team has barcoded 3.6M specimens - but has been restricted to fungi, plants, and animals. With help, he says the project can expand to register bacterial and Archaea genomes as well.
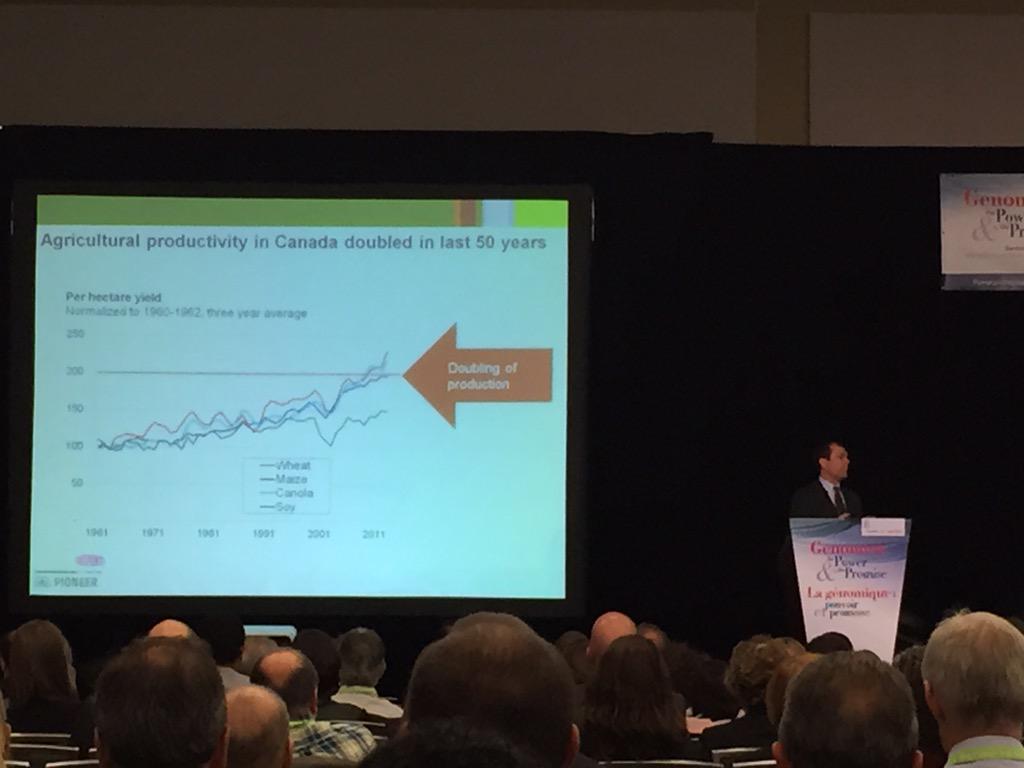 When we talk about climate change, we (lay people) forget to factor in population growth. According to Doane Chilcoat of DuPont Pioneer, we're looking at a population of 9B people by 2050. We'll need to feed all of those people, but that's not exactly easy. Canada hasn't increased its farmland since the 1970s, so what are we to do? On top of that, a warmer climate means less water availability (even if rainfall doesn't change), and it also means more bugs to damage the crops. How do we reduce this pressure on agriculture in the future? Dr. Chilcoat suggests expanding and intensifying production, and increasing the efficiency of agriculture by reducing waste, improving cultural practices, and introducing better genetics. By using genomics technologies, researchers can predict and identify key traits that are best adapted for our future climate.
When we talk about climate change, we (lay people) forget to factor in population growth. According to Doane Chilcoat of DuPont Pioneer, we're looking at a population of 9B people by 2050. We'll need to feed all of those people, but that's not exactly easy. Canada hasn't increased its farmland since the 1970s, so what are we to do? On top of that, a warmer climate means less water availability (even if rainfall doesn't change), and it also means more bugs to damage the crops. How do we reduce this pressure on agriculture in the future? Dr. Chilcoat suggests expanding and intensifying production, and increasing the efficiency of agriculture by reducing waste, improving cultural practices, and introducing better genetics. By using genomics technologies, researchers can predict and identify key traits that are best adapted for our future climate. This is the same type of work that Sally Aitken from the University of British Columbia has been working on. She uses genomics technology to identify traits in spruce and pine trees that will enable them to better adapt to their changing climate, in a project called AdapTree. This information is used to make policy recommendations in managing forest land that is regulated by provincial governments. Because the life-span of trees is so long, they can experience a huge range of climate change in their lifetimes. Dr. Aitken stresses that if we can plant trees that are pre-adapted to future climates, we can address the problem of maladaptation and productivity loss in forestry. In her work, she has found that the frequencies of many SNPs were correlated with the landscape environment for specific populations of trees. Her future work will validate the candidate adaptive SNPs, compare the adaptive phenotypes and genotypes of natural v. selectively bred populations, and use AdapTree SNP arrays for pilot genomic selection in tree breeding programs.
At the evening gala, we heard from Sharon Moalem, who spoke about the value of the rare and why exceptional genetic conditions matter. It's hard to find funding for research when there are only a handful of people in the world with that disease, so what are we to do? Well, Dr. Moalem started by asking us what the greatest killer in world history was. If you guessed the plague or the flu, you'd be wrong. It was cyanobacteria, who created oxygen and started the Great Oxidation, which killed off all the previous life that used iron as a its main element. In fact, most of the bacteria in our bodies are actually mining us for iron, and the "friendly" bacteria are those that don't deplete our iron reserves. Dr. Moalem spent his graduate level research studying hemochromatosis, a disorder in which the body sequesters iron to the point where the organs rust from excess iron. By mapping the frequency of C282Y, he found that the frequency of this gene increased following the plague in Western Europe, making it far more common - with a frequency of about 30% - after that time than it was before. In doing this research, Dr. Moalem found a new class of antibiotics - siderocillins - as well as some great new drugs that lower cholesterol. So why are rare diseases important to study? Because they define the limits of human physiology, and they can tell us so much more about other, more common diseases.
Tomorrow we'll be wrapping up the conference with a session on genomics solutions for the non-renewable energy sector. I'll be tweeting again from my own account and the Genome Canada account. Anyone wishing to follow along should look under the #powerofgenomics feed.
Thanks again for sharing the day Dominique. As I'm no longer working in molecular biology, and on the West Coast it's great to have access to some insight.
ReplyDeleteI'm really interested in what you reported about the microbiome, since we tend to forget that we have fewer of our own cells in our bodies that "foreign" ones, and they must make a huge impact.
As someone who suffered a psychotic break brought on by life stress and now with a diagnosis of bipolar disorder I was intrigued how mental illness can be prevented by policy...unless we stop people from going to work and having children - or even having interactions with other people, this could be tricky :-) Joking aside, t does illustrate the importance of awareness and the affects of circumstances on mental health.
I'm also a former crop scientist, involved with crop protection, so that aspect of genomics is of interest...the references from UBC about trees resistant to climate change were inspiring.
Looking forward to your tweets tomorrow. I may even get up early to catch the first session :-)